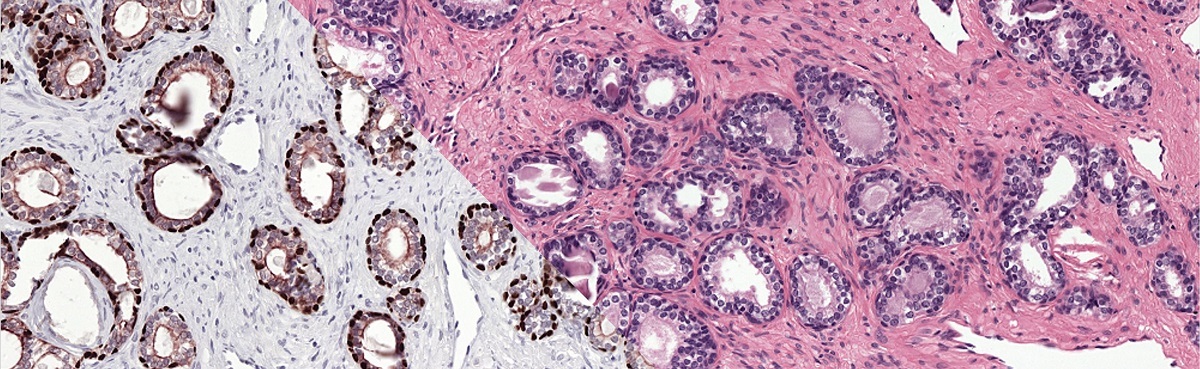
As part of our publication on epithelium segmentation using deep learning and immunohistochemistry we published our dataset on Zenodo: the PESO dataset. The dataset is free to use under the BY-NC-SA Creative Commons license.

The reference standard of the PESO set was made using immunohistochemistry, resulting in a precise and accurate ground truth.
Contents of the dataset
The PESO dataset consists 102 whole-slide images, split in to in a training and test part. The total dataset is around 140GB. For the training part, the reference standard is included. This set consists of:
- 62 whole-slide images, exported at a pixel resolution of 0.48mu/pixels.
- 62 Raw color deconvolution masks containing the P63&CK8/18 channel of the color deconvolution operation. These masks mark all regions that are stained by either P63 or CK8/18 in the IHC version of the slides.
- 25 color deconvolution masks (N=25) on which manual annotations have been made. Within these regions, stain and other artifacts have been removed.
- 62 training masks (N=62) that have been used to train the main network of our paper. These masks are generated by a trained U-Net on the corresponding IHC slides.
The test set consists of:
- 40 whole-slide images, exported at a pixel resolution of 0.48mu/pixels.
- 40 xml files containing a total of 160 annotations of regions that are used in the original evaluation.
- 160 png files of 2500x2500 pixels, exported at a pixel resolution of 0.48mu/pixels. Each png file corresponds to one test region.
- 160 padded png files of 3500x3500 pixels of the same test regions.
- A mapping file (csv) describing whether a test region contains cancer or only benign tissue.

Example from the PESO dataset. The dataset consists of full whole-slide images.

Example of the ground truth segmentation of the epithelium tissue.
Using the data
All slides are saved in TIFF format and can be opened with a viewer such as ASAP. To use the images in Python we can use the python binding of ASAP. To do so, make sure that the <ASAP install directory>/bin is in your PYTHONPATH. Then use the following snippet to extract a patch:
# Import the ASAP module
import multiresolutionimageinterface as mir
# Create a new reader to open files
reader = mir.MultiResolutionImageReader()
image = reader.open('path to image file')
# Get a patch at x=1000, y=12000 (coordinates relative to level 0) at level 2
level = 1
patch = image.getUCharPatch(10000, 12000, 1000, 1000, level)
More info / questions?
Background information on the dataset can be found in the paper. For questions please use the contact form.
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
AI for diagnosis of prostate cancer: the PANDA challenge
Artificial intelligence (AI) for prostate cancer analysis is ready for clinical implementation, shows a global programming competition, the PANDA challenge.
Improve prostate cancer diagnosis: participate in the PANDA Gleason grading challenge
Can you build a deep learning model that can accurately grade protate biopsies? Participate in the PANDA challenge
The potential of AI in medicine: AI-assistance improves prostate cancer grading
In a completely new study we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.