With 1.1 million new diagnoses every year, prostate cancer (PCa) is second most common cancer among males worldwide. The biopsy Gleason grading system is the strongest prognostic marker for prostate cancer but suffers from significant inter-observer variability, limiting its usefulness for individual patients.
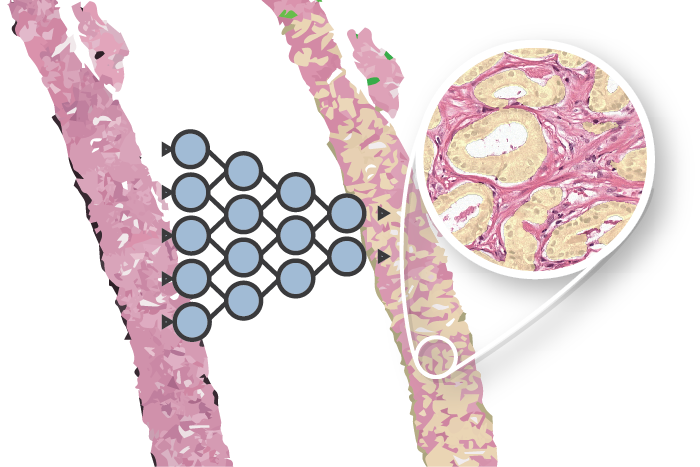
Automated deep learning systems have shown promise in accurately grading prostate cancer1,2. Several studies have shown that these systems can achieve pathologist-level performance, including my own research on automated Gleason grading. These developments have the potential to assisting pathologists in their diagnostic process. However, A large multi-center evaluation on diagnostic data is still missing.
Do you want to help us to improve prostate cancer diagnosis?
The PANDA challenge
Researchers and staff from Radboud Universiy Medical Center, Karolinska Institutet, Tampere University and Kaggle are organizing the PANDA challenge: Prostate cANcer graDe Assessment using the Gleason grading system. With this challenge we aim to set the next step in automated Gleason grading for prostate cancer.
For this challenge, we are releasing all training data of two major studies1,2 on automated Gleason grading, both previously published in Lancet Oncology. The training set contains almost 11.000 prostate biopsies, with slide level labels and label masks. All submitted methods are evaluated on a private set of around 500 biopsies, split between public and private leaderboard.
The top three teams are eligble for a cash prize. The total amount of prize money available for the challenge is $25.000. Additionally, we will invite five teams to present their method the challenge workshop at MICCAI 2020.
After the challenge, the data can also be used for other research. We will release the data under a CC BY-NC-SA license. We ask challenge participants to respect an embargo until the challenge paper has been published. After the embargo is lifted, participants are free to publish their own results.
How to participate
To participate in the challenge, please go to the competition page on Kaggle for more information. All challenge data can be downloaded from the Kaggle platform.
Please signup up before July 15th to participate in the challenge. The final submission must be submitted July 22th the latest.
MICCAI 2020 Workshop

The PANDA challenge is part of the MICCAI 2020 conference. During MICCAI, we will host a workshop on the challenge and the overall results. Up to five teams will be invited to present their method. We will select teams for a presentations mostly based on ranking, but can deviate to diversify in the methods presented.
Series on my PhD in Computational Pathology
This post is part of a series related to my PhD project on prostate cancer, deep learning and computational pathology. In my research I developed AI algorithms to diagnose prostate cancer. Interested in the rest of my research? The three latest post are shown below. For all the posts, you can find all posts tagged with research.
Latest post related to my research
AI for diagnosis of prostate cancer: the PANDA challenge
Artificial intelligence (AI) for prostate cancer analysis is ready for clinical implementation, shows a global programming competition, the PANDA challenge.
The potential of AI in medicine: AI-assistance improves prostate cancer grading
In a completely new study we investigated the possible benefits of an AI system for pathologists. Instead of focussing on pathologist-versus-AI, we instead look at potential pathologist-AI synergy.
Epithelium segmentation using deep learning and immunohistochemistry
We developed a new deep learning method to segment epithelial tissue in digitized hematoxylin and eosin (H&E) stained prostatectomy slides using immunohistochemistry (IHC) as reference standard.
Deep learning posts
Sometimes I write a blog post on deep learning or related techniques. The latest posts are shown below. Interested in reading all my blog posts? You can read them on my tech blog.
Simple and efficient data augmentations using the Tensorfow tf.Data and Dataset API
The tf.data API of Tensorflow is a great way to build a pipeline for sending data to the GPU. In this post I give a few examples of augmentations and how to implement them using this API.
Getting started with GANs Part 2: Colorful MNIST
In this post we build upon part 1 of 'Getting started with generative adversarial networks' and work with RGB data instead of monochrome. We apply a simple technique to map MNIST images to RGB.
Getting started with generative adversarial networks (GAN)
Generative Adversarial Networks (GANs) are one of the hot topics within Deep Learning right now and are applied to various tasks. In this post I'll walk you through the first steps of building your own adversarial network with Keras and MNIST.
References
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. The Lancet Oncology 21, 233-241, Read online (2020). ↩︎ ↩︎
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. The Lancet Oncology 21, 222-232, Read online (2020). ↩︎ ↩︎
